ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
UC3 : Application of Meta-PanGenomic concept for tracking antibioresistance in metagenomic samples

Members
Coordinator :
David Vallenet (LABGeM, Genoscope, CEA) & Nicolas Pons (MetaGenoPolis, INRAE)
Team members :
Claudine Médigue (LABGeM, CNRS, Genoscope ; IFB)
Mathieu Almeida (MetaGenoPolis, INRAE)
Emmanuelle Le Chatelier (MetaGenoPolis, INRAE)
Florian Plaza Onate (MetaGenoPolis, INRAE)
Guillaume Gautreau (MaIAGE, INRAE)
Objective and outcome
This use case aims to demonstrate the potential application of Meta-PanGenomic concept for tracking at the species and strain levels associations between antimicrobial resistance and pathogenicity in the context of human microbiota.
Outcome: Guidelines and reproducible workflows for applying Meta-PanGenomic approach in the ABRomics-PF.
Reconstruction of metagenome-assembled genomes (MAGs) has recently become a common task for microbiome studies in particular to compare genetic content of species across metagenomic samples. However, it has been shown that the gene content captured in a MAG can highly differ from the one obtained by the genome sequencing of the corresponding isolate (Meziti, 2021; Maguire, 2020). Missing genes in MAGs are commonly harbored by genomic regions with atypical tetranucleotide frequency that make difficult the application of assembly and binning tools to reconstruct them. These atypical genomic regions correspond frequently to mobile genetic elements that may contain virulence factors and antimicrobial resistance genes (ARG) (Oliveira, 2017). Long read technologies (like Oxford Nanopore or Pacific Bioscience) could be a game-changer in the next few years and will be useful in particular for discovery purposes rather than quantitative estimation of known variations.
Therefore, in order to better characterize and quantify the dissemination of antimicrobial resistance and its association with potential pathogenicity, we propose for this application an innovative approach combining the use of metagenomic data and pangenome graph structure (Gautreau, 2020). For a given microbial species, a pangenome graph combines the genomic content of all the microbial strains (optionally enriched with high-quality MAGs and metagenomic long read assemblies) where nodes represent the gene families and edges their genomic neighborhood. As said, antimicrobial resistance and virulence genes are associated with specific genetic contexts including pathogenicity islands and/or mobile genetic elements. Therefore, the gene families of the pangenome graph are partitioned in core and accessory elements in order to identify, in association with their functional annotation, specific regions of interest which are variable in the pangenome. In the context of the identification of a pathogenic bacterial strain from an environmental or clinical sample, a direct mapping of metagenomics reads on the nodes of the species pangenome graph will highlight relevant regions containing ARG and/or virulence factors. In the case of complex samples with several strains of the same species, the distribution of the mapping abundances along the pangenome graph can be analyzed with adapted signal deconvolution methods to unravel strain composition in a sample with their relative abundances and specific variable genomic regions. To summarize, the described approach could be named Meta-PanGenomic as it uses a pangenome database queried by the metagenomic reads to deeply characterize and quantify antimicrobial-resistant strains.
The development of the Meta-PanGenomic graph method will be conducted in strong interaction with WP3 of ABRomics. The strategies already developed by the LABGeM partner using PPanGGOLiN software (Gautreau, 2020) will be a starting point. Aims of the use-case will be to iterate specific methodological needs with WP3 and to develop, benchmark and validate the approach. Interactions with WP2 will be required in order to address metagenomic data processing with standardized pipelines of ABRomics and pangenome annotation for antimicrobial resistance genes, virulence factors and mobile genetic elements.
This use-case will be applied on well-chosen metagenomic datasets in order to demonstrate the technical feasibility and the scientific interest in the context of antibioresistance.
To address this issue, we propose to use the recent EcoZUR cohort (Peña-Gonzalez, 2019) focused on the role of E. coli on diarrhea in children. This study makes available metagenomic samples of children with or without diarrhea. Causal agents (E. coli) have been isolated and sequenced from each sample of diarrheic children. Here, known causal agents will be tracked by mapping metagenomic samples on a specific pangenome graph aggregating reference genomes of E. coli isolates including those sequenced in the EcoZUR study. This dataset will permit to benchmark and validate the development of the Meta-PanGenomic approach. In particular, we will explore the impact of adding variable-quality MAGs to pangenome graphs for the detection of variable regions. To note that this dataset could be completed by the German outbreak study (Loman, 2013).
We propose to address a potential application of pangenomic graph analysis in clinical metagenomic by using a published study related to the effect of antibiotics intake on the gut microbiota stability (Palleja, 2018). This study provides metagenomic samples of gut microbiota of 12 healthy men over a 6-month period following a 4-day intervention with a cocktail of 3 last-resort antibiotics (meropenem, gentamicin and vancomycin). Pangenome graphs will be analysed for gut species enriched quickly after antibiotics intake in order to detect resistant bacterial strains and potentially associated with microbiota virulence enrichment. Using longitudinal metagenomic samples, we will also explore the long-term impact of antibiotics intake on the microbiota changes and potential antimicrobial resistance gene acquisitions and emergence of pathogen resistant bacterial strains.
The French Gut Project and Million Microbiomes of Human Project (MMHP) (in which MetaGenoPolis partner is coordinator and member, respectively) will characterize the microbiota diversity across large populations. These initiatives will make open access every year the metagenomic data accompanied with an anonymized minimal set of metadata describing each volunteer. For ABRomics consortium, access to such large cohorts will permit to explore the resistome diversity and its evolution according to age, geography, or certain clinical status. We propose to investigate these datasets using the pangenomic graph concept in order to map the ‘world-wide’ antimicrobial resistance diversity of E. coli and the other ‘most wanted bacteria’ of the WHO.
Using Meta-PanGenome graph methods together with ABRomics platform services, the use-case will address scientific issues related to antibioresistance in link with the human microbiome:
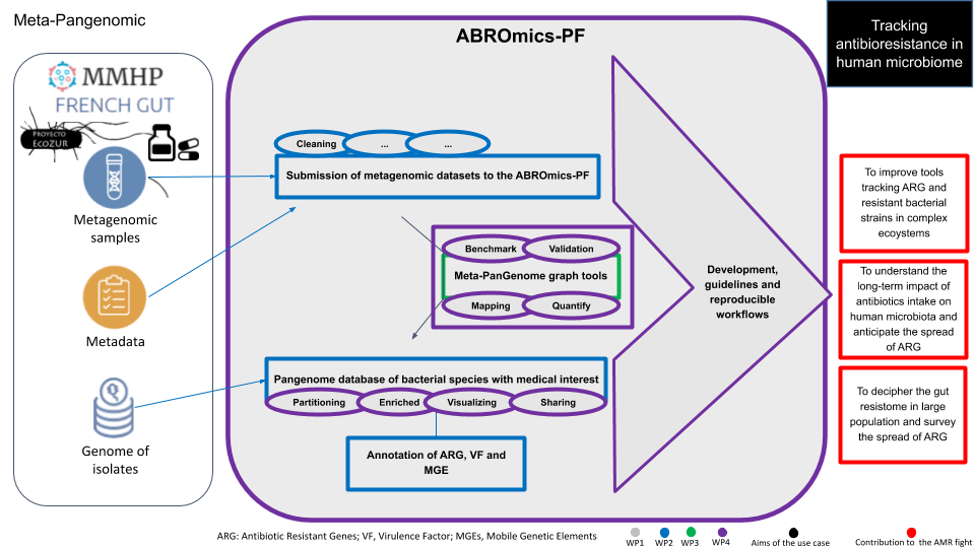
Meta-Pangenomic use-case diagram describing roles and interactions with WP in the development and integration of the Meta-PanGenomic concept in the ABRomics-PF and its application in ARG tracking in human microbiome.