- By Nam Phan Van Song
- ABRomics
ABRomics team presents an ongoing work on knowledge graph and ontology at the SWAT4HCLS conference
ABRomics team members, Brieuc Quemener and Alban Gaignard, are working on the data integration objective of work-package (WP3) at the BiRD Platform in Nantes. They were invited to give a talk to Semantic Web Application and Tools for Health Care and Life Sciences (SWAT4HCLS) international conference, which was held on the 24 to 27 February in Barcelona. Brieuc presented an ongoing work on the ABRomics knowledge graph, which is based on selected existing ABR (AntiBioResistance) ontologies.
For now the ABRomics platform relies on a Postgres database to store all the data of the platform. User data, projects, analysis results and metadata of imported fastq files all end up in this relational database. This approach has been proven resilient to storing massive amounts of data but also opens the door to difficulties when it comes to sharing this data to third party databases as our database schema is unique to the ABRomics platform.
On the other hand, knowledge graphs can overcome these data interoperability issues. The analysis data can be transformed into nodes and links forming a knowledge graph. Then, with the use of consensus terms listed in popular ABR ontologies (such as ARO), the nodes of the graph can be well annotated. The graph structure allows anyone to query the graph natively using SPARQL queries, while adherence to ABR ontologies provides the terms that will guide the crafting of queries while ensuring that our knowledge graph can be queried alongside other ABR knowledge graphs.
Using the SOSA ontology to represent multimodal data
However, ABR relevant ontologies such as ARO, GO, NCIT, ENVO, FOODON do not provide a way to store the analytics data produced by the ABRomics workflows. Therefore, although these ontologies are relevant for annotating data at a later stage, we had to find another ontology that provides a structure capable of storing multimodal data.
The short article, accepted at the SWAT4HCLS conference, focuses on the use of the SOSA ontology as a blueprint to store ABRomics analysis data into a knowledge graph. Initially used to represent sensor networks, the SOSA ontology structure is close to the way the analysis process occurs on ABRomics. A sample is analysed using a workflow which produces results of various types: classes of antibiotics detected, taxonomy of the analysed strain, etc.
To learn more
In fact, a sample can be considered as a sosa:FeatureOfInterest. On this sample, a workflow will be triggered, which corresponds to a sosa:Procedure. The workflow produces a lot of sosa:Observations on the sample. A sosa:Observation can be made on different sosa:ObservableProperty, like the name of an antibiotic resistance gene found in the sample or the coverage of the gene. Every sosa:Observation also contains the raw value of the observation (the name of the gene for instance).
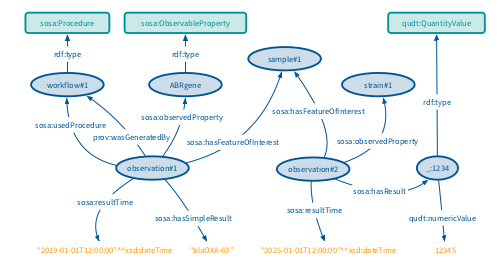
Example RDF instances for the data analysis results.
The SOSA ontology also integrates the notion of time and space,enabling data to be queried using multiple modalities. It allows to perform queries such as : “get all the resistance genes that were found in France between the 10/02/2024 and the 10/02/2025”
We built the knowledge graph by extracting public Acinetobacter baumannii analysis data from the ABRomics platform. Then we parsed the data into a knowledge graph structured with the SOSA ontology. Afterward, we bind the terms to other ontologies such as ARO, NCIT and GO to improve data interoperability. To support queries on geographical data, we relied on wikidata and its terms.
Conclusion
As the result, this knowledge graph allowed us to successfully answer one of the first biological question of interest : “What are the most represented A. baumannii antibiotic resistance genes in a specific geographical region of interest ?”
A demo of the knowledge graph capacities is available here. It allows tusers to play with the first two competency questions and modify them slightly to have a better grasp on how queries are structured.
The SWAT4HCLS conference was a great opportunity to share our ongoing works on the ontology search process and the use of SOSA to store data for which this ontology was not initially designed.. This will also be an opportunity to get feedback from experts all around the world and discuss with people having similar objectives !
For more information:
Brieuc Quemeneur, Audrey Bihouée, Samuel Chaffron, Claudine Médigue, Hervé Ménager, et al.. A
multi-modal and temporal antibiotic resistance knowledge graph. SWAT4HCLS 2025 – 16th International SWAT4HCLS conference : Semantic Web Applications and Tools for Health Care and Life
Sciences, Scientifika, Feb 2025, Barcelone, Spain. https://hal.science/hal-04976716
All the source code of the knowledge graph and the web demo are available on github: https://github.com/Phloemus/ABRomics-KG





